
BIT scoring system of Python_MadMapper_V112_RECBIT.py script for RILs (recombinant inbred lines) is similar to scoring system for alignments of protein or DNA sequences. Genotype data for particular marker for the set of RILs can be considered as a sequence (string). Comparison of two or more genotype datasets can be considered as a comparison of two or more strings (sequences). As for protein/DNA sequence alignment approach there is a scoring matrix. For recombination inbred lines (RILs) scoring matrix may look like:
#################################################################
# +-------+ #
# | BIT | #
# SCORING SYSTEM: | | #
# | REC | #
# +-------+ #
# #
# . +-------+-------+-------+-------+-------+-------+ #
# . | | | | | | | #
# . | A | B | C | D | H | - | #
# .| | | | | | | #
# +-------*-------+-------+-------+-------+-------+-------+ #
# | | . 6 | -6 | -4 | 4 | -2 | 0 | #
# | A | | | | | | | #
# | | 0 .| 1 | 1 | 0 | 0.5 | 0 | #
# +-------+-------*-------+-------+-------+-------+-------+ #
# | | -6 | . 6 | 4 | -4 | -2 | 0 | #
# | B | | | | | | | #
# | | 1 | 0 .| 0 | 1 | 0.5 | 0 | #
# +-------+-------+-------*-------+-------+-------+-------+ #
# | | -4 | 4 | . 4 | -4 | 0 | 0 | #
# | C | | | | | | | #
# | | 1 | 0 | 0 .| 1 | 0 | 0 | #
# +-------+-------+-------+-------*-------+-------+-------+ #
# | | 4 | -4 | -4 | . 4 | 0 | 0 | #
# | D | | | | | | | #
# | | 0 | 1 | 1 | 0 .| 0 | 0 | #
# +-------+-------+-------+-------+-------*-------+-------+ #
# | | -2 | -2 | 0 | 0 | . 2 | 0 | #
# | H | | | | | | | #
# | | 0.5 | 0.5 | 0 | 0 | 0 .| 0 | #
# +-------+-------+-------+-------+-------+-------*-------+ #
# | | 0 | 0 | 0 | 0 | 0 | . 0 | #
# | - | | | | | | | #
# | | 0 | 0 | 0 | 0 | 0 | 0 .| #
# +-------+-------+-------+-------+-------+-------+-------*. #
# #
# #
# NOTES: #
# C - NOT A ( H or B ) #
# D - NOT B ( H or A ) #
# H - A and B #
# #
#################################################################
Values in this particular scoring matrix were determined empirically after several "try to see what happen" experiments. These values were adjusted to get good and reproducible results for the set of RILs in range of 100 - 200 individuals of generation 8 or higher. However, most likely, they will work with smaller or large datasets. Examples how this scoring works you can see below:
#################################################################
# #
# EXAMPLES OF SCORING: #
# #
# #
# POSITIVE LINKAGE: #
# #
# AAAAAAAAAAAAAAAAAAAA BIT SCORE = 6*20 = 120 #
# AAAAAAAAAAAAAAAAAAAA REC SCORE = 0 (0.0) #
# .. #
# AAAAAAAAAAAAAAAAAAAA BIT SCORE = 6*18 - 6*2 = 96 #
# AAAAAAAAAAAAAAAAAABB REC SCORE = 2 (2/20 = 0.1) #
# #
# AAAAAAAAAABBBBBBBBBB BIT SCORE = 6*10 + 6*10 = 120 #
# AAAAAAAAAABBBBBBBBBB REC SCORE = 0 (0.0) #
# .. #
# AAAAAAAAABABBBBBBBBB BIT SCORE = 6*18 - 6*2 = 96 #
# AAAAAAAAAABBBBBBBBBB REC SCORE = 2 (2/20 = 0.1) #
# #
# #
# NO LINKAGE: #
# .......... #
# AAAAAAAAAAAAAAAAAAAA BIT SCORE = 6*10 - 6*10 = 0 #
# AAAAAAAAAABBBBBBBBBB REC SCORE = 10 (10/20 = 0.5) #
# . . . . . . . . . . #
# BBBAABBAAAAAAABAABBB BIT SCORE = 6*10 - 6*10 = 0 #
# BABBAABBABABABBBAABA REC SCORE = 10 (10/20 = 0.5) #
# #
# #
# NEGATIVE LINKAGE: #
# .................. #
# AAAAAAAAAAAAAAAAAAAA BIT SCORE = 6*2 - 6*18 = -96 #
# AABBBBBBBBBBBBBBBBBB REC SCORE = 18 (18/20 = 0.9) #
# .................. #
# ABABABABABABABABABAB BIT SCORE = 6*2 - 6*18 = -96 #
# ABBABABABABABABABABA REC SCORE = 18 (18/20 = 0.9) #
# #
# #
#################################################################
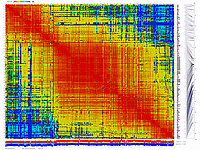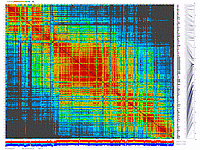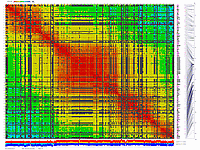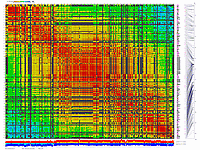
We have applied this approach to real dataset of RILs for Arabidopsis. Results of scoring and clustering you can find here.
email to: akozik@atgc.org Alexander Kozik
last modified July 27 2004